Extended Data Figure 5: Characterization of sequence and structure at m1A sites, and requirement of the TRMT6/TRMT61A machinery for its catalysis in the cytosol.

Extended Data Figure 3: Calibration of quantum-noise-limited Faraday rotation probing of atomic spins.

Extended Data Figure 6: Marginalized posterior distributions of the radial-velocity model parameters.

Extended Data Figure 2: Relationship between net N mineralization and rainfall over the study periods.
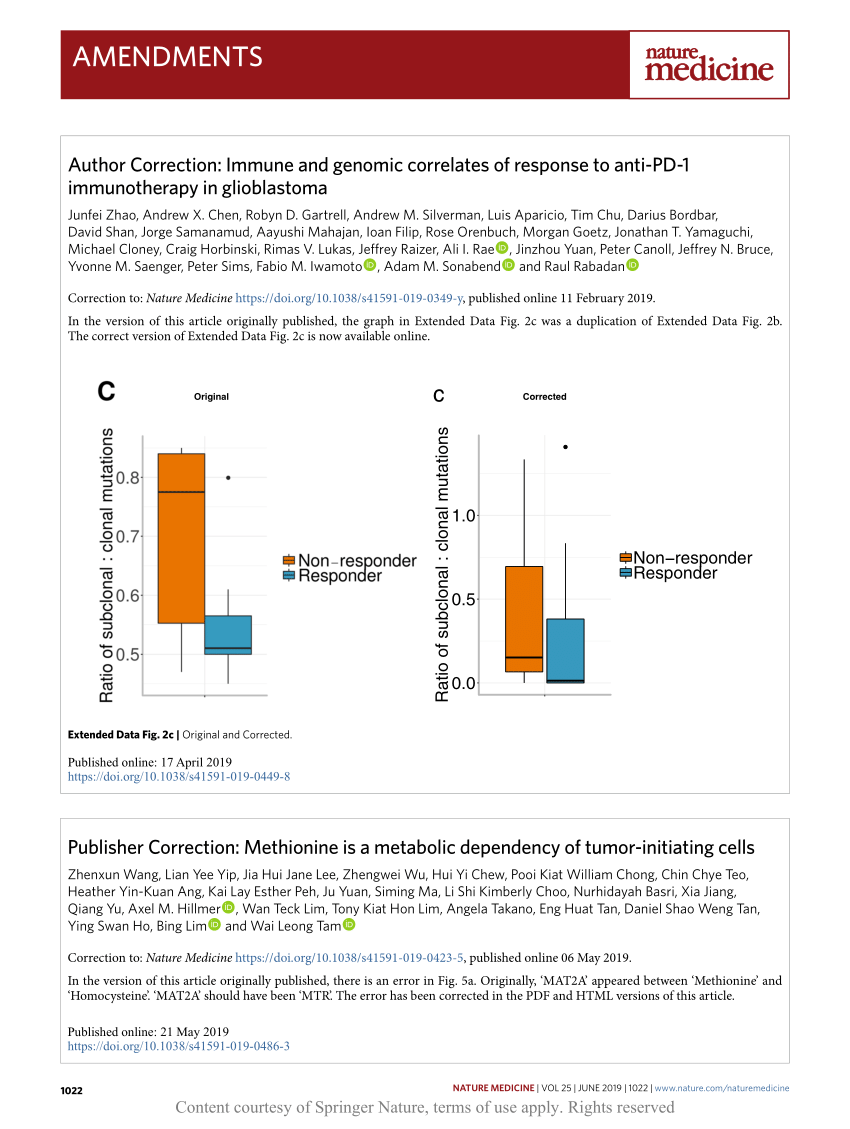
Author Correction: Immune and genomic correlates of response to anti-PD-1 immunotherapy in glioblastoma | Request PDF